Background
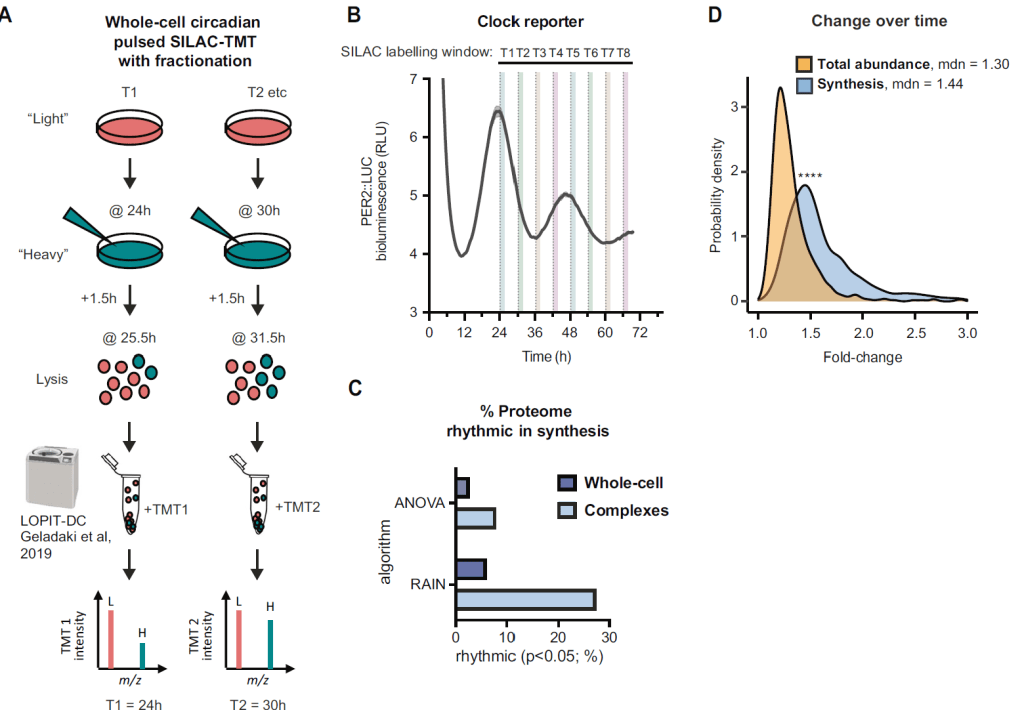
I did my PhD and subsequently a short postdoc in the lab of John O’Neill at the MRC Laboratory of Molecular Biology in Cambridge, UK. My research there focused on circadian rhythms and protein homeostasis, and I ended up doing a lot of proteomics analysis and looking at macromolecular protein complexes. I worked on a variety of other topics, incl. a biophysics project on water potential and cellular macromolecular condensation, where I developed a novel method for phosphoproteomics analysis, and that ended up published in Nature. (so now I can be done with academia, right?)
After MRC LMB, I worked at RxCelerate Ltd, a small biotech CRO, as a Proteomics Scientist delivering the ProQuant(R) platform and doing internal R&D. It was great to experience the biotech world and I learnt a bunch more proteomics & coding too. I am now enjoying being back in academia (possibly an unpopular opinion?) at the UCL Cancer Institute, where I am working within the Proteomics Research Translational Technology Platform and the Bioinformatics Hub as a Research Fellow to help other scientists, students, and clinicians to design, deliver and analyse proteomics as well as multi-omics data.
Topics of interest
-omics (post-translational) & data integration
To date, I have been mostly working with proteomics and phosphoproteomics data, but also with other biological data and -omics. I am particularly interested in integrating various datasets (-omics, knowledge databases, structure information) to gain new biological insights.
More
Although I don’t have a formal computational background, I started doing proteomics analysis in 2020(ish), and continue to develop my skills in that, as well as more general bioinformatics.
In my PhD and first postdoc, I’ve employed TMT, dynamic SILAC, and phosphoproteomics (among other methods) to study temporal regulation and protein complex formation. I have performed other types of -omics analysis, including RNAseq, metabolomics, and multi-omics, and always found it crucial to both apply appropriate statistical modelling approaches, as well as robustly explore -omics data in the biological context, e.g. integrating with public datasets, protein network and pathway analysis, gene ontology, etc. At RxCelerate/ProQuant, I worked with the company’s clients to design and analyse proteomics studies, taking data from raw formats to biological insight, as well as led on internal method development, incl. developing bioinformatics QC procedures and setting up new workflows, e.g. for DIA.
Cell & cancer biology
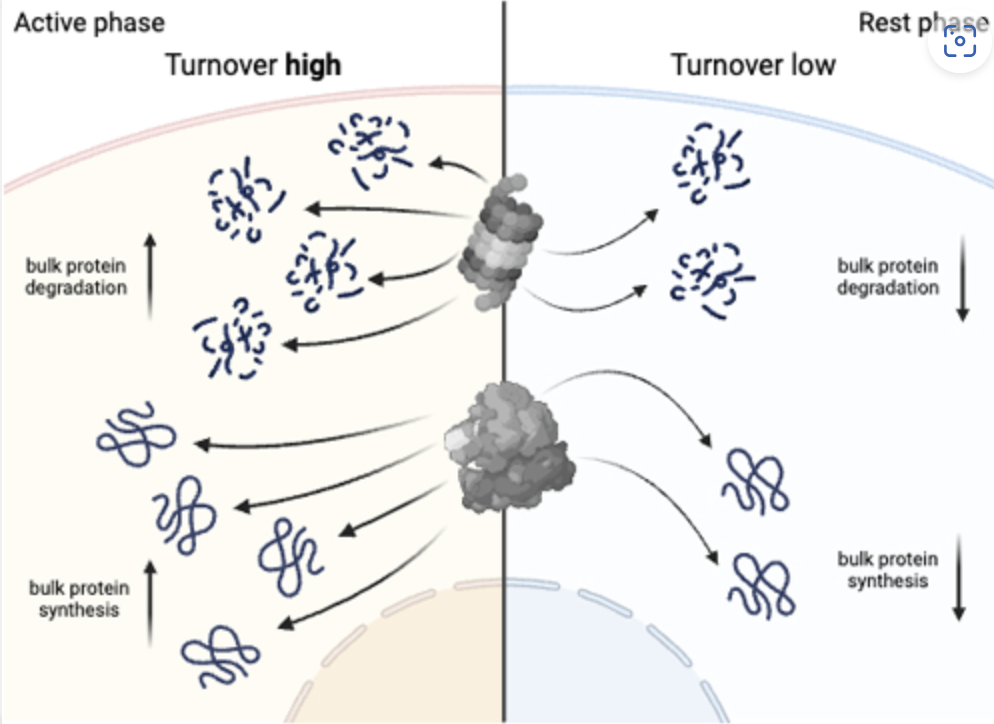
I am fascinated by questions in cell biology, including but not limited to: post-translational regulation, circadian rhythms, protein homeostasis, PTMs, protein internal disorder. I am starting to work in cancer biology as part of my new job at UCL.

Cellular agriculture
Cell ag is a field I am very familiar with through community-building work, but I have not worked in it as a scientist directly – yet. If you have a project that you think could use my expertise (e.g. in proteomics or other bioinformatics) – I’d be very keen to explore collaboration options!
Publications
E. Seinkmane, et al., Circadian regulation of macromolecular complex turnover and proteome renewal. EMBO J (2024)
J. Watson, E. Seinkmane, et al., Rapid changes in protein condensation buffer intracellular water against osmotic and thermal challenge, Nature (2024)
D. Wong, E. Seinkmane, et al., CRYPTOCHROMES promote daily protein homeostasis. EMBO J. (2022).
A. Stangherlin, E. Seinkmane, J. O’Neill, Understanding circadian regulation of mammalian cell function, protein homeostasis, and metabolism. Curr. Opin. Syst. Biol. (2021).
A. Stangherlin, […], E. Seinkmane, […], et al., Compensatory ion transport buffers daily protein rhythms to regulate osmotic balance and cellular physiology. Nat. Commun. (2021).
M. Putker, D. Wong, E. Seinkmane, et al., CRYPTOCHROMES confer robustness, not rhythmicity, to circadian timekeeping. EMBO J. (2021)
K. Wilson, […], E. Seinkmane, et al., Glycans modify mesenchymal stem cell differentiation to impact on the function of resulting osteoblasts. J. Cell Sci. (2018)
N. Hoyle, E. Seinkmane, et al., Circadian actin dynamics drive rhythmic fibroblast mobilization during wound healing. Sci. Transl. Med. (2017)
N. Rzechorzek, […], E.Seinkmane, et al., Circadian clocks in human cerebral organoids. bioRxiv (2024)